Malus is a genus with over 35 species naturally distributed across the temperate northern hemisphere, from East Asia and Europe to North America. This genus includes the domesticated apple (Malus domestica) and its wild relatives. New research reveals the evolutionary relationships among Malus species and how their genomes have evolved over the past nearly 60 million years.
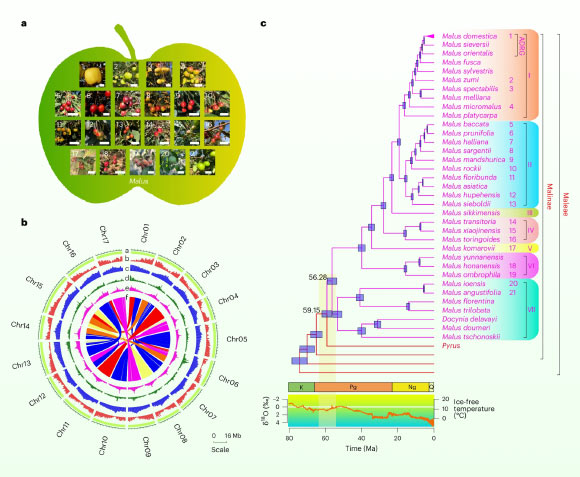
The Malus evolutionary landscape based on phylogenomics. Image credit: Li et al., doi: 10.1038/s41588-025-02166-6.
“There are roughly 35 species in the genus Malus, but despite the importance of apple as a fruit crop, there hasn’t been extensive study of how this group’s genomes have evolved,” said Penn State Professor Hong Ma.
“In this study, we were able to do a deep dive into the genomes of Malus, establish an apple family tree, document events like whole-genome duplications and hybridizations between species, and find regions of the genome associated with specific traits, like resistance to apple scab disease.”
Professor Ma and colleagues newly sequenced and assembled the genomes of 30 members of the Malus genus, including the domesticated golden delicious apple variety.
Of the 30 species, 20 are diploid, meaning that they have two copies of each chromosome, like humans, and 10 are polyploid, having three or four copies of each chromosome, likely due to a relatively recent hybridizations of diploid and other relatives in Malus.
By comparing the sequence of nearly 1,000 genes from each species, the researchers built a family tree of the genus and then used biogeographical analysis to trace its origin to about 56 million years ago in Asia.
“The evolutionary history of the genus is quite complex, with numerous examples of hybridization between species and a shared whole-genome duplication event that make comparisons difficult,” Professor Ma said.
“Having high-quality genomes for such a large number of the species in the genus and understanding the relationships among them allowed us to dig deeper into how the genus has evolved.”
To further analyze the history and evolution of the Malus genomes, the scientists examined the 30 sequenced genomes in an analytical approach called pan-genomics.
This approach involved comprehensive comparison for both shared, or conserved, genes and other sequences, such as transposons — sometimes called jumping genes for their ability to move in the genome — across the 30 genomes, as well as genes that are only present in subsets of the genomes.
Pan-genomic analyses combine the genomic information from a closely related group to understand evolutionary conservation and divergences and were greatly facilitated by the pan-genome graph tool.
“The use of the pan-genome of 30 species was powerful for detecting structural variation, as well as gene duplications and rearrangements, among the species that might be missed by comparisons of only a few genomes,” Professor Ma said.
“In this case, one of the uncovered structural variants allowed us to pinpoint the genome segment associated with resistance to apple scab, a fungal disease that impacts apples worldwide.”
The authors also developed a pan-genome analysis tool to help find evidence of selective sweeps, a process where a beneficial trait rapidly increases in frequency in a population.
Using this method, they identified a genome region responsible for cold and disease resistance in wild Malus species that also may be related to unpleasant taste in fruit.
“It’s possible that in the efforts to produce the best tasting fruit, there was an inadvertent reduction of the hardiness of domesticated apples,” Professor Ma said.
“Understanding the structural variations in the Malus genomes, the relationships among the species and their history of hybridization using pan-genome analysis could help guide future breeding efforts so that the beneficial traits for good taste and disease-resistant can both be retained in apples.”
The findings were published in the journal Nature Genetics.
_____
W. Li et al. Pan-genome analysis reveals the evolution and diversity of Malus. Nat Genet, published online April 16, 2025; doi: 10.1038/s41588-025-02166-6










